This is the as.matrix() method for projection objects (returned by
project(), possibly as elements of a list). It extracts the projected
parameter draws and returns them as a matrix. In case of different (i.e.,
nonconstant) weights for the projected draws, see
as_draws_matrix.projection() for a better solution.
Usage
# S3 method for class 'projection'
as.matrix(x, nm_scheme = NULL, allow_nonconst_wdraws_prj = FALSE, ...)Arguments
- x
An object of class
projection(returned byproject(), possibly as elements of alist).- nm_scheme
The naming scheme for the columns of the output matrix. Either
NULL,"rstanarm", or"brms", whereNULLchooses"rstanarm"or"brms"based on the class of the reference model fit (and uses"rstanarm"if the reference model fit is of an unknown class).- allow_nonconst_wdraws_prj
A single logical value indicating whether to allow projected draws with different (i.e., nonconstant) weights (
TRUE) or not (FALSE). CAUTION: Expert use only because if set toTRUE, the weights of the projected draws are stored in an attributewdraws_prjand handling this attribute requires special care (e.g., when subsetting the returned matrix).- ...
Currently ignored.
Value
An \(S_{\mathrm{prj}} \times Q\) matrix of projected
draws, with \(S_{\mathrm{prj}}\) denoting the number of projected
draws and \(Q\) the number of parameters. If allow_nonconst_wdraws_prj
is set to TRUE, the weights of the projected draws are stored in an
attribute wdraws_prj. (If allow_nonconst_wdraws_prj is FALSE,
projected draws with nonconstant weights cause an error.)
Details
In case of the augmented-data projection for a multilevel submodel
of a brms::categorical() reference model, the multilevel parameters (and
therefore also their names) slightly differ from those in the brms
reference model fit (see section "Augmented-data projection" in
extend_family()'s documentation).
Examples
# Data:
dat_gauss <- data.frame(y = df_gaussian$y, df_gaussian$x)
# The `stanreg` fit which will be used as the reference model (with small
# values for `chains` and `iter`, but only for technical reasons in this
# example; this is not recommended in general):
fit <- rstanarm::stan_glm(
y ~ X1 + X2 + X3 + X4 + X5, family = gaussian(), data = dat_gauss,
QR = TRUE, chains = 2, iter = 500, refresh = 0, seed = 9876
)
# Projection onto an arbitrary combination of predictor terms (with a small
# value for `ndraws`, but only for the sake of speed in this example; this
# is not recommended in general):
prj <- project(fit, predictor_terms = c("X1", "X3", "X5"), ndraws = 21,
seed = 9182)
# Applying the as.matrix() generic to the output of project() dispatches to
# the projpred::as.matrix.projection() method:
prj_mat <- as.matrix(prj)
# Since the draws have all the same weight here, we can treat them like
# ordinary MCMC draws, e.g., we can summarize them using the `posterior`
# package:
if (requireNamespace("posterior", quietly = TRUE)) {
print(posterior::summarize_draws(
posterior::as_draws_matrix(prj_mat),
"median", "mad", function(x) quantile(x, probs = c(0.025, 0.975))
))
}
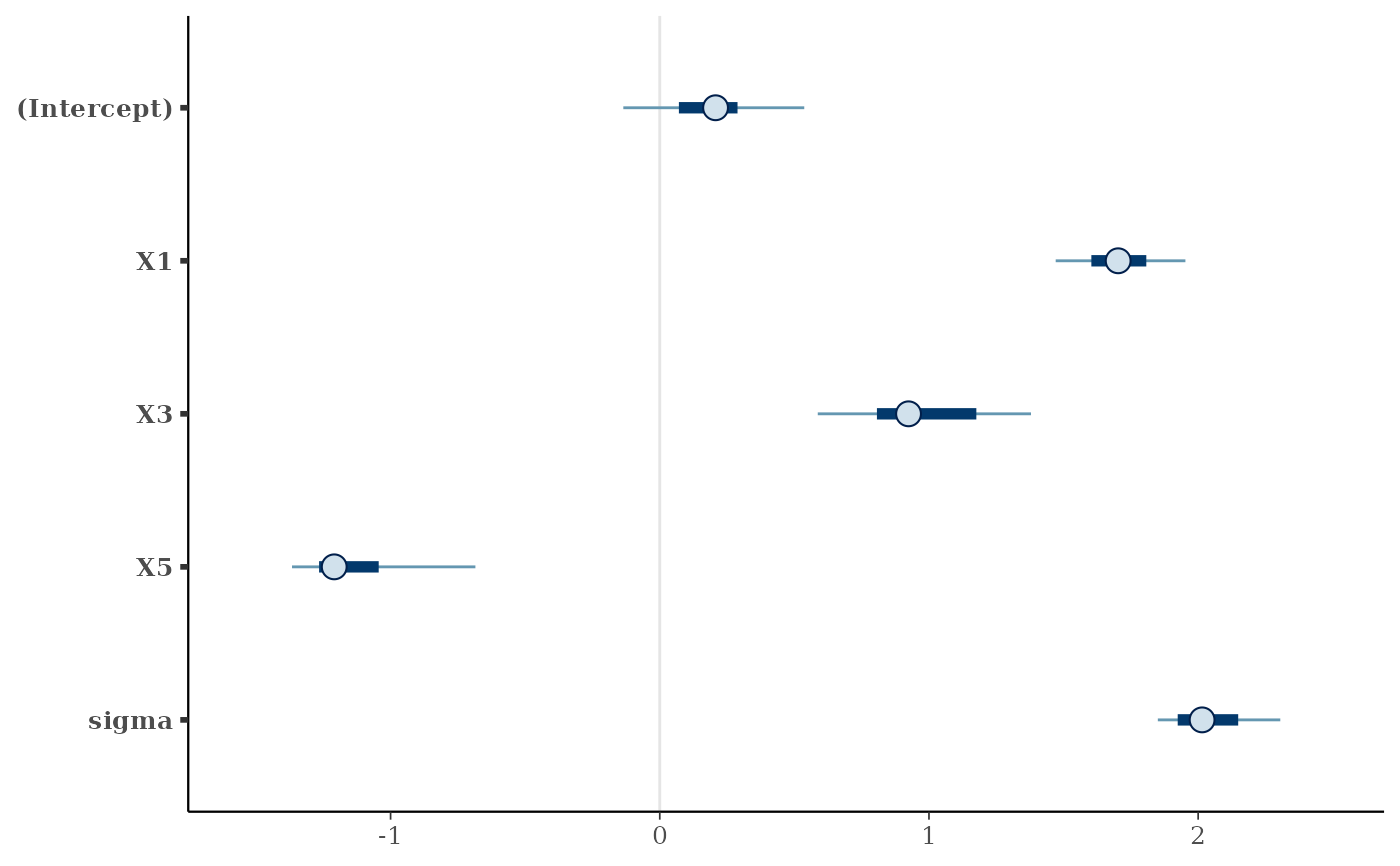
#> # A tibble: 5 × 5
#> variable median mad `2.5%` `97.5%`
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 0.207 0.202 -0.269 0.594
#> 2 X1 1.70 0.155 1.40 1.97
#> 3 X3 0.924 0.266 0.560 1.45
#> 4 X5 -1.21 0.111 -1.53 -0.602
#> 5 sigma 2.01 0.183 1.83 2.35
# Or visualize them using the `bayesplot` package:
if (requireNamespace("bayesplot", quietly = TRUE)) {
print(bayesplot::mcmc_intervals(prj_mat))
}