Implementation of Pareto smoothed importance sampling (PSIS), a method for stabilizing importance ratios. The version of PSIS implemented here corresponds to the algorithm presented in Vehtari, Simpson, Gelman, Yao, and Gabry (2024). For PSIS diagnostics see the pareto-k-diagnostic page.
Usage
psis(log_ratios, ...)
# S3 method for class 'array'
psis(log_ratios, ..., r_eff = 1, cores = getOption("mc.cores", 1))
# S3 method for class 'matrix'
psis(log_ratios, ..., r_eff = 1, cores = getOption("mc.cores", 1))
# Default S3 method
psis(log_ratios, ..., r_eff = 1)
is.psis(x)
is.sis(x)
is.tis(x)Arguments
- log_ratios
An array, matrix, or vector of importance ratios on the log scale (for PSIS-LOO these are negative log-likelihood values). See the Methods (by class) section below for a detailed description of how to specify the inputs for each method.
- ...
Arguments passed on to the various methods.
- r_eff
Vector of relative effective sample size estimates containing one element per observation. The values provided should be the relative effective sample sizes of
1/exp(log_ratios)(i.e.,1/ratios). This is related to the relative efficiency of estimating the normalizing term in self-normalizing importance sampling. Ifr_effis not provided then the reported PSIS effective sample sizes and Monte Carlo error estimates can be over-optimistic. If the posterior draws are (near) independent thenr_eff=1can be used.r_effhas to be a scalar (same value is used for all observations) or a vector with length equal to the number of observations. The default value is 1. See therelative_eff()helper function for computingr_eff.- cores
The number of cores to use for parallelization. This defaults to the option
mc.coreswhich can be set for an entire R session byoptions(mc.cores = NUMBER). The old optionloo.coresis now deprecated but will be given precedence overmc.coresuntilloo.coresis removed in a future release. As of version 2.0.0 the default is now 1 core ifmc.coresis not set, but we recommend using as many (or close to as many) cores as possible.Note for Windows 10 users: it is strongly recommended to avoid using the
.Rprofilefile to setmc.cores(using thecoresargument or settingmc.coresinteractively or in a script is fine).
- x
For
is.psis(), an object to check.
Value
The psis() methods return an object of class "psis",
which is a named list with the following components:
log_weightsVector or matrix of smoothed (and truncated) but unnormalized log weights. To get normalized weights use the
weights()method provided for objects of class"psis".diagnosticsA named list containing two vectors:
pareto_k: Estimates of the shape parameter \(k\) of the generalized Pareto distribution. See the pareto-k-diagnostic page for details.n_eff: PSIS effective sample size estimates.
Objects of class "psis" also have the following attributes:
norm_const_logVector of precomputed values of
colLogSumExps(log_weights)that are used internally by theweightsmethod to normalize the log weights.tail_lenVector of tail lengths used for fitting the generalized Pareto distribution.
r_effIf specified, the user's
r_effargument.dimsInteger vector of length 2 containing
S(posterior sample size) andN(number of observations).methodMethod used for importance sampling, here
psis.
Methods (by class)
psis(array): An \(I\) by \(C\) by \(N\) array, where \(I\) is the number of MCMC iterations per chain, \(C\) is the number of chains, and \(N\) is the number of data points.psis(matrix): An \(S\) by \(N\) matrix, where \(S\) is the size of the posterior sample (with all chains merged) and \(N\) is the number of data points.psis(default): A vector of length \(S\) (posterior sample size).
References
Vehtari, A., Gelman, A., and Gabry, J. (2017). Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC. Statistics and Computing. 27(5), 1413–1432. doi:10.1007/s11222-016-9696-4 (journal version, preprint arXiv:1507.04544).
Vehtari, A., Simpson, D., Gelman, A., Yao, Y., and Gabry, J. (2024). Pareto smoothed importance sampling. Journal of Machine Learning Research, 25(72):1-58. PDF
See also
loo()for approximate LOO-CV using PSIS.pareto-k-diagnostic for PSIS diagnostics.
The loo package vignettes for demonstrations.
The FAQ page on the loo website for answers to frequently asked questions.
Examples
log_ratios <- -1 * example_loglik_array()
r_eff <- relative_eff(exp(-log_ratios))
psis_result <- psis(log_ratios, r_eff = r_eff)
str(psis_result)
#> List of 2
#> $ log_weights: num [1:1000, 1:32] 2.37 2.12 2.24 2.41 2.25 ...
#> $ diagnostics:List of 3
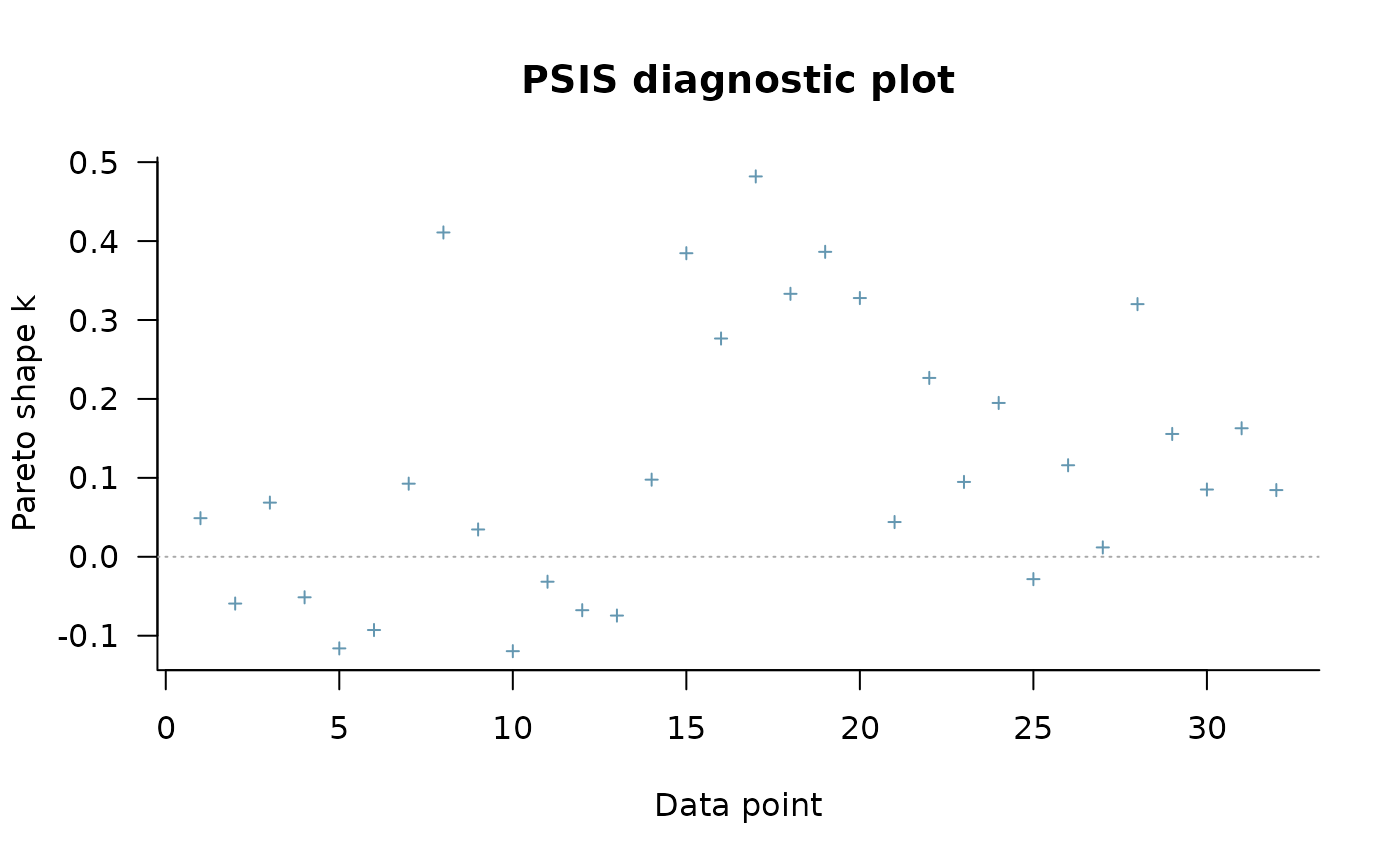
#> ..$ pareto_k: num [1:32] 0.0489 -0.0593 0.0686 -0.0513 -0.1161 ...
#> ..$ n_eff : num [1:32] 909 937 938 901 907 ...
#> ..$ r_eff : num [1:32] 0.942 0.954 0.977 0.919 0.923 ...
#> - attr(*, "norm_const_log")= num [1:32] 9.28 9.04 9.25 9.09 9 ...
#> - attr(*, "tail_len")= num [1:32] 98 98 96 99 99 101 99 100 102 98 ...
#> - attr(*, "r_eff")= num [1:32] 0.942 0.954 0.977 0.919 0.923 ...
#> - attr(*, "dims")= int [1:2] 1000 32
#> - attr(*, "method")= chr "psis"
#> - attr(*, "class")= chr [1:3] "psis" "importance_sampling" "list"
plot(psis_result)
 # extract smoothed weights
lw <- weights(psis_result) # default args are log=TRUE, normalize=TRUE
ulw <- weights(psis_result, normalize=FALSE) # unnormalized log-weights
w <- weights(psis_result, log=FALSE) # normalized weights (not log-weights)
uw <- weights(psis_result, log=FALSE, normalize = FALSE) # unnormalized weights
# extract smoothed weights
lw <- weights(psis_result) # default args are log=TRUE, normalize=TRUE
ulw <- weights(psis_result, normalize=FALSE) # unnormalized log-weights
w <- weights(psis_result, log=FALSE) # normalized weights (not log-weights)
uw <- weights(psis_result, log=FALSE, normalize = FALSE) # unnormalized weights