Bayesian generalized linear additive models with optional group-specific terms via Stan
Source:R/stan_gamm4.R
stan_gamm4.RdBayesian inference for GAMMs with flexible priors.
Usage
stan_gamm4(
formula,
random = NULL,
family = gaussian(),
data,
weights = NULL,
subset = NULL,
na.action,
knots = NULL,
drop.unused.levels = TRUE,
...,
prior = default_prior_coef(family),
prior_intercept = default_prior_intercept(family),
prior_smooth = exponential(autoscale = FALSE),
prior_aux = exponential(autoscale = TRUE),
prior_covariance = decov(),
prior_PD = FALSE,
algorithm = c("sampling", "meanfield", "fullrank"),
adapt_delta = NULL,
QR = FALSE,
sparse = FALSE
)
plot_nonlinear(
x,
smooths,
...,
prob = 0.9,
facet_args = list(),
alpha = 1,
size = 0.75
)Arguments
- formula, random, family, data, knots, drop.unused.levels
Same as for
gamm4. We strongly advise against omitting thedataargument. Unlessdatais specified (and is a data frame) many post-estimation functions (includingupdate,loo,kfold) are not guaranteed to work properly.- subset, weights, na.action
Same as
glm, but rarely specified.- ...
Further arguments passed to
sampling(e.g.iter,chains,cores, etc.) or tovb(ifalgorithmis"meanfield"or"fullrank").- prior
The prior distribution for the (non-hierarchical) regression coefficients.
The default priors are described in the vignette Prior Distributions for rstanarm Models. If not using the default,
priorshould be a call to one of the various functions provided by rstanarm for specifying priors. The subset of these functions that can be used for the prior on the coefficients can be grouped into several "families":Family Functions Student t family normal,student_t,cauchyHierarchical shrinkage family hs,hs_plusLaplace family laplace,lassoProduct normal family product_normalSee the priors help page for details on the families and how to specify the arguments for all of the functions in the table above. To omit a prior —i.e., to use a flat (improper) uniform prior—
priorcan be set toNULL, although this is rarely a good idea.Note: Unless
QR=TRUE, ifprioris from the Student t family or Laplace family, and if theautoscaleargument to the function used to specify the prior (e.g.normal) is left at its default and recommended value ofTRUE, then the default or user-specified prior scale(s) may be adjusted internally based on the scales of the predictors. See the priors help page and the Prior Distributions vignette for details on the rescaling and theprior_summaryfunction for a summary of the priors used for a particular model.- prior_intercept
The prior distribution for the intercept (after centering all predictors, see note below).
The default prior is described in the vignette Prior Distributions for rstanarm Models. If not using the default,
prior_interceptcan be a call tonormal,student_torcauchy. See the priors help page for details on these functions. To omit a prior on the intercept —i.e., to use a flat (improper) uniform prior—prior_interceptcan be set toNULL.Note: If using a dense representation of the design matrix —i.e., if the
sparseargument is left at its default value ofFALSE— then the prior distribution for the intercept is set so it applies to the value when all predictors are centered (you don't need to manually center them). This is explained further in [Prior Distributions for rstanarm Models](https://mc-stan.org/rstanarm/articles/priors.html) If you prefer to specify a prior on the intercept without the predictors being auto-centered, then you have to omit the intercept from theformulaand include a column of ones as a predictor, in which case some element ofpriorspecifies the prior on it, rather thanprior_intercept. Regardless of howprior_interceptis specified, the reported estimates of the intercept always correspond to a parameterization without centered predictors (i.e., same as inglm).- prior_smooth
The prior distribution for the hyperparameters in GAMs, with lower values yielding less flexible smooth functions.
prior_smoothcan be a call toexponentialto use an exponential distribution, ornormal,student_torcauchy, which results in a half-normal, half-t, or half-Cauchy prior. Seepriorsfor details on these functions. To omit a prior —i.e., to use a flat (improper) uniform prior— setprior_smoothtoNULL. The number of hyperparameters depends on the model specification but a scalar prior will be recylced as necessary to the appropriate length.- prior_aux
The prior distribution for the "auxiliary" parameter (if applicable). The "auxiliary" parameter refers to a different parameter depending on the
family. For Gaussian modelsprior_auxcontrols"sigma", the error standard deviation. For negative binomial modelsprior_auxcontrols"reciprocal_dispersion", which is similar to the"size"parameter ofrnbinom: smaller values of"reciprocal_dispersion"correspond to greater dispersion. For gamma modelsprior_auxsets the prior on to the"shape"parameter (see e.g.,rgamma), and for inverse-Gaussian models it is the so-called"lambda"parameter (which is essentially the reciprocal of a scale parameter). Binomial and Poisson models do not have auxiliary parameters.The default prior is described in the vignette Prior Distributions for rstanarm Models. If not using the default,
prior_auxcan be a call toexponentialto use an exponential distribution, ornormal,student_torcauchy, which results in a half-normal, half-t, or half-Cauchy prior. Seepriorsfor details on these functions. To omit a prior —i.e., to use a flat (improper) uniform prior— setprior_auxtoNULL.- prior_covariance
Cannot be
NULL; seedecovfor more information about the default arguments.- prior_PD
A logical scalar (defaulting to
FALSE) indicating whether to draw from the prior predictive distribution instead of conditioning on the outcome.- algorithm
A string (possibly abbreviated) indicating the estimation approach to use. Can be
"sampling"for MCMC (the default),"optimizing"for optimization,"meanfield"for variational inference with independent normal distributions, or"fullrank"for variational inference with a multivariate normal distribution. Seerstanarm-packagefor more details on the estimation algorithms. NOTE: not all fitting functions support all four algorithms.- adapt_delta
Only relevant if
algorithm="sampling". See the adapt_delta help page for details.- QR
A logical scalar defaulting to
FALSE, but ifTRUEapplies a scaledqrdecomposition to the design matrix. The transformation does not change the likelihood of the data but is recommended for computational reasons when there are multiple predictors. See the QR-argument documentation page for details on how rstanarm does the transformation and important information about how to interpret the prior distributions of the model parameters when usingQR=TRUE.- sparse
A logical scalar (defaulting to
FALSE) indicating whether to use a sparse representation of the design (X) matrix. IfTRUE, the the design matrix is not centered (since that would destroy the sparsity) and likewise it is not possible to specify bothQR = TRUEandsparse = TRUE. Depending on how many zeros there are in the design matrix, settingsparse = TRUEmay make the code run faster and can consume much less RAM.- x
An object produced by
stan_gamm4.- smooths
An optional character vector specifying a subset of the smooth functions specified in the call to
stan_gamm4. The default is include all smooth terms.- prob
For univarite smooths, a scalar between 0 and 1 governing the width of the uncertainty interval.
- facet_args
An optional named list of arguments passed to
facet_wrap(other than thefacetsargument).- alpha, size
For univariate smooths, passed to
geom_ribbon. For bivariate smooths,size/2is passed togeom_contour.
Details
The stan_gamm4 function is similar in syntax to
gamm4 in the gamm4 package. But rather than performing
(restricted) maximum likelihood estimation with the lme4 package,
the stan_gamm4 function utilizes MCMC to perform Bayesian
estimation. The Bayesian model adds priors on the common regression
coefficients (in the same way as stan_glm), priors on the
standard deviations of the smooth terms, and a prior on the decomposition
of the covariance matrices of any group-specific parameters (as in
stan_glmer). Estimating these models via MCMC avoids
the optimization issues that often crop up with GAMMs and provides better
estimates for the uncertainty in the parameter estimates.
See gamm4 for more information about the model
specicification and priors for more information about the
priors on the main coefficients. The formula should include at least
one smooth term, which can be specified in any way that is supported by the
jagam function in the mgcv package. The
prior_smooth argument should be used to specify a prior on the unknown
standard deviations that govern how smooth the smooth function is. The
prior_covariance argument can be used to specify the prior on the
components of the covariance matrix for any (optional) group-specific terms.
The gamm4 function in the gamm4 package uses
group-specific terms to implement the departure from linearity in the smooth
terms, but that is not the case for stan_gamm4 where the group-specific
terms are exactly the same as in stan_glmer.
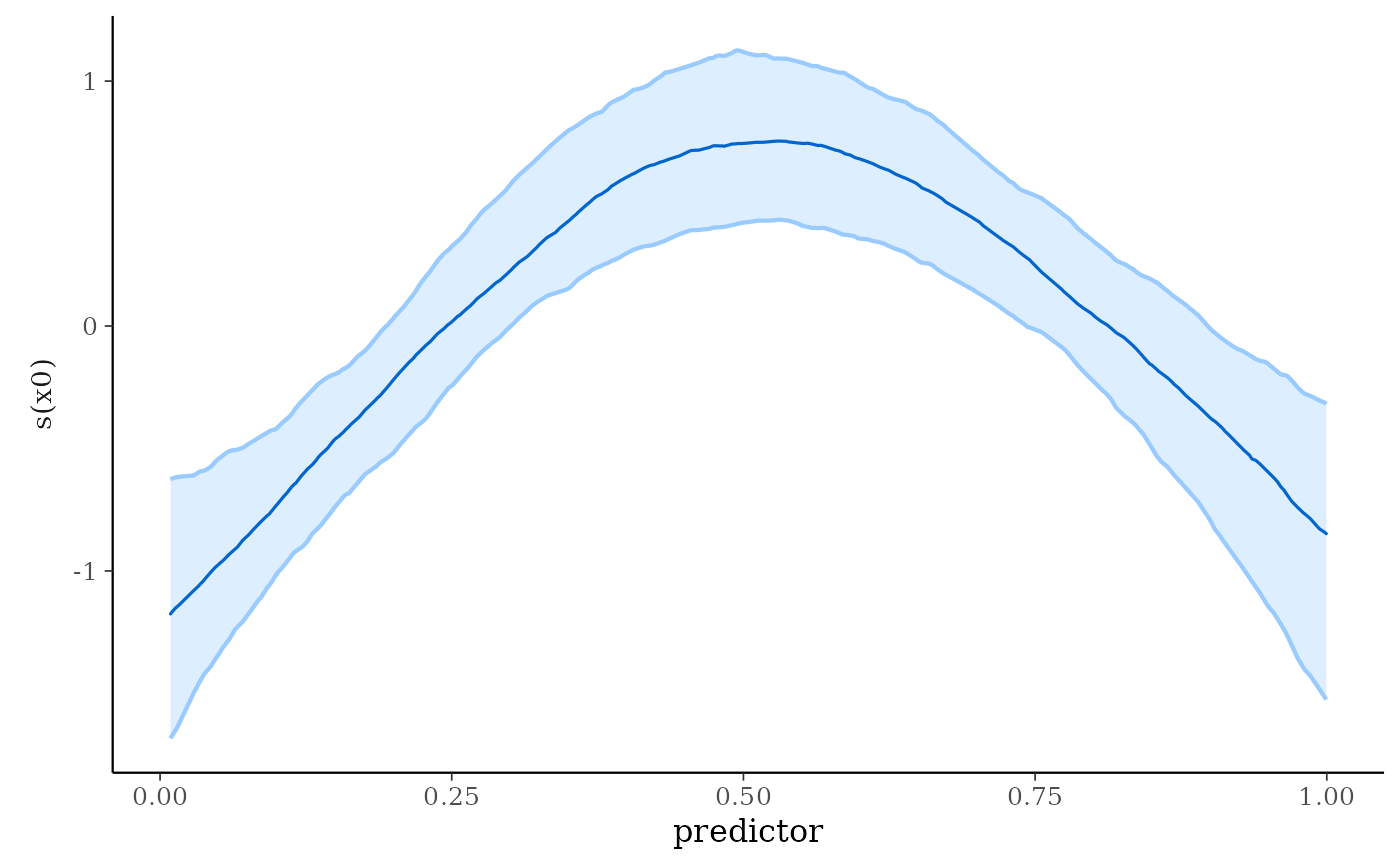
The plot_nonlinear function creates a ggplot object with one facet for
each smooth function specified in the call to stan_gamm4 in the case
where all smooths are univariate. A subset of the smooth functions can be
specified using the smooths argument, which is necessary to plot a
bivariate smooth or to exclude the bivariate smooth and plot the univariate
ones. In the bivariate case, a plot is produced using
geom_contour. In the univariate case, the resulting
plot is conceptually similar to plot.gam except the
outer lines here demark the edges of posterior uncertainty intervals
(credible intervals) rather than confidence intervals and the inner line
is the posterior median of the function rather than the function implied
by a point estimate. To change the colors used in the plot see
color_scheme_set.
References
Crainiceanu, C., Ruppert D., and Wand, M. (2005). Bayesian analysis for penalized spline regression using WinBUGS. Journal of Statistical Software. 14(14), 1–22. https://www.jstatsoft.org/article/view/v014i14
See also
stanreg-methods and
gamm4.
The vignette for stan_glmer, which also discusses
stan_gamm4. https://mc-stan.org/rstanarm/articles/
Examples
if (.Platform$OS.type != "windows" || .Platform$r_arch != "i386") {
# from example(gamm4, package = "gamm4"), prefixing gamm4() call with stan_
# \donttest{
dat <- mgcv::gamSim(1, n = 400, scale = 2) ## simulate 4 term additive truth
## Now add 20 level random effect `fac'...
dat$fac <- fac <- as.factor(sample(1:20, 400, replace = TRUE))
dat$y <- dat$y + model.matrix(~ fac - 1) %*% rnorm(20) * .5
br <- stan_gamm4(y ~ s(x0) + x1 + s(x2), data = dat, random = ~ (1 | fac),
chains = 1, iter = 500) # for example speed
print(br)
plot_nonlinear(br)
plot_nonlinear(br, smooths = "s(x0)", alpha = 2/3)
# }
}
#> Gu & Wahba 4 term additive model
#>
#> SAMPLING FOR MODEL 'continuous' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 7.9e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.79 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 500 [ 0%] (Warmup)
#> Chain 1: Iteration: 50 / 500 [ 10%] (Warmup)
#> Chain 1: Iteration: 100 / 500 [ 20%] (Warmup)
#> Chain 1: Iteration: 150 / 500 [ 30%] (Warmup)
#> Chain 1: Iteration: 200 / 500 [ 40%] (Warmup)
#> Chain 1: Iteration: 250 / 500 [ 50%] (Warmup)
#> Chain 1: Iteration: 251 / 500 [ 50%] (Sampling)
#> Chain 1: Iteration: 300 / 500 [ 60%] (Sampling)
#> Chain 1: Iteration: 350 / 500 [ 70%] (Sampling)
#> Chain 1: Iteration: 400 / 500 [ 80%] (Sampling)
#> Chain 1: Iteration: 450 / 500 [ 90%] (Sampling)
#> Chain 1: Iteration: 500 / 500 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 2.475 seconds (Warm-up)
#> Chain 1: 1.245 seconds (Sampling)
#> Chain 1: 3.72 seconds (Total)
#> Chain 1:
#> Warning: There were 6 divergent transitions after warmup. See
#> https://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup
#> to find out why this is a problem and how to eliminate them.
#> Warning: Examine the pairs() plot to diagnose sampling problems
#> Warning: The largest R-hat is 1.05, indicating chains have not mixed.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#r-hat
#> Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#bulk-ess
#> Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#tail-ess
#> stan_gamm4
#> family: gaussian [identity]
#> formula: y ~ s(x0) + x1 + s(x2)
#> observations: 400
#> ------
#> Median MAD_SD
#> (Intercept) 5.2 0.2
#> x1 5.6 0.4
#> s(x0).1 -0.2 1.6
#> s(x0).2 -0.1 2.1
#> s(x0).3 0.1 2.0
#> s(x0).4 0.2 1.6
#> s(x0).5 -0.1 1.7
#> s(x0).6 -2.6 1.4
#> s(x0).7 0.2 0.8
#> s(x0).8 -1.8 1.6
#> s(x0).9 0.0 0.8
#> s(x2).1 -31.8 13.3
#> s(x2).2 -21.1 9.9
#> s(x2).3 23.3 8.1
#> s(x2).4 18.1 5.2
#> s(x2).5 -29.0 4.3
#> s(x2).6 13.1 2.1
#> s(x2).7 12.4 2.1
#> s(x2).8 -11.0 4.9
#> s(x2).9 6.1 9.0
#>
#> Auxiliary parameter(s):
#> Median MAD_SD
#> sigma 2.1 0.1
#>
#> Smoothing terms:
#> Median MAD_SD
#> smooth_sd[s(x0)1] 2.1 0.8
#> smooth_sd[s(x0)2] 1.2 1.2
#> smooth_sd[s(x2)1] 16.7 3.1
#> smooth_sd[s(x2)2] 5.2 4.9
#>
#> Error terms:
#> Groups Name Std.Dev.
#> fac (Intercept) 0.75
#> Residual 2.15
#> Num. levels: fac 20
#>
#> ------
#> * For help interpreting the printed output see ?print.stanreg
#> * For info on the priors used see ?prior_summary.stanreg